×
![]()
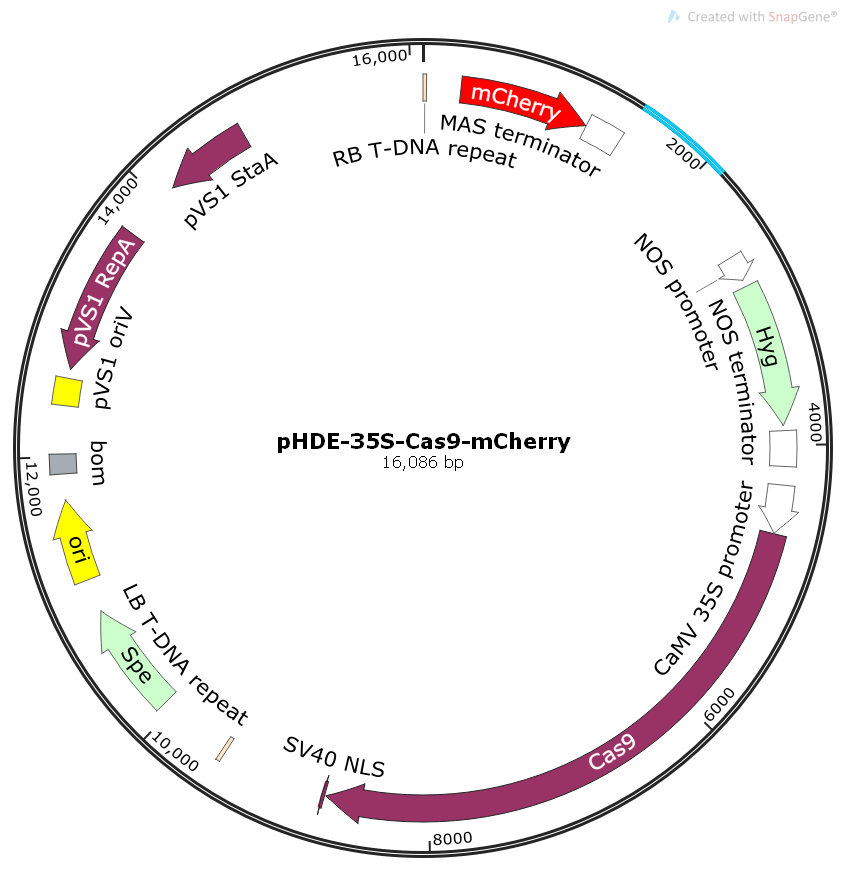
- 规格:20μl质粒
- 启动子:CaMV 35S promoter
- 复制子:pUC ori,pVS1 oriV
- 终止子:3A terminator
- 质粒分类:植物系列质粒;植物编辑质粒;植物Cas9质粒
- 质粒标签:C-SV40 NLS
- 原核抗性:壮观霉素Spe
- 筛选标记:潮霉素Hygro,红色荧光蛋白mCherry
- 克隆菌株:Stbl3等大肠杆菌
- 表达宿主:拟南芥等植物
- 诱导方式:无需诱导,组成型表达
- 5测序引物:35S:GACGCACAATCCCACTATCC
- 3测序引物:3A-R:CCATTTGCATTTTGATGTCC
- 质粒简介: pHDE-35S-Cas9-mCherry质粒可以在植物中表达Cas9酶和mCherry红色荧光蛋白,可视化基因编辑操作。For CRISPR/Cas9 mediated gene editing in Arabidopsis; provides a visual screen for Cas9-free plants.其参考文献是An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing.
- 质粒图谱1: png/1-1P12G33F11S.png
质粒序列: 点击查看序列LOCUS Exported 16086 bp ds-DNA circular SYN 27-JAN-2018FEATURES Location/Qualifierssource 1..16086/organism="synthetic DNA construct"/mol_type="other DNA"misc_feature 1..25/label=RB T-DNA repeat/note="right border repeat from nopaline C58 T-DNA"CDS 271..1197/codon_start=1/label=mCherry/translation="MMSGIVLFVTCHSFPTHNPDNNVRASLSLHTHSCMHAFLHVIAMQISFLTYKYKPTLHYTLHSNQNKKTYTNSKTVPTMDNMAIIKEFMRFKVHMEGSVNGHEFEIEGEGEGRPYEGTQTAKLKVTKGGPLPFAWDILSPQFMYGSKAYVKHPADIPDYLKLSFPEGFKWERVMNFEDGGVVTVTQDSSLQDGEFIYKVKLRGTNFPSDGPVMQKKTMGWEASSERMYPEDGALKGEIKQRLKLKDGGHYDAEVKTTYKAKKPVQLPGAYNVNIKLDITSHNEDYTIVEQYERAEGRHSTGGMDELYK"terminator 1203..1455/label=MAS terminator/note="mannopine synthase terminator"promoter 2602..2785/function="NOS promoter"/label=NOS promoter/note="nopaline synthase promoter"CDS 2837..3862/codon_start=1/label=Hyg/translation="MKKPELTATSVEKFLIEKFDSVSDLMQLSEGEESRAFSFDIGGRGYVLRVNSCADGFYKDRYVYRHFASAALPIPEVLDIGEFSESLTYCISRRAQGVTLQDLPETELPAVLQPVAEAMDAIAAADLSQTSGFGPFGPQGIGQYTTWRDFICAIADPHVYHWQTVMDDTVSASVAQALDELMLWAEDCPEVRHLVHADFGSNNVLTDNGRITAVIDWSEAMFGDSQYEVANIFFWRPWLACMEQQTRYFERRHPELAGSPRLRAYMLRIGLDQLYQSLVDGNFDDAAWAQGRCDAIVRSGAGTVGRTQIARRSAAVWTDGCVEVLADSGNRRPSTRPGSWR"terminator 3900..4152/label=NOS terminator/note="nopaline synthase terminator and poly(A) signal"promoter 4285..4630/label=CaMV 35S promoter/note="strong constitutive promoter from cauliflower mosaicvirus"CDS 4635..8738/codon_start=1/product="Cas9 (Csn1) endonuclease from the Streptococcuspyogenes Type II CRISPR/Cas system"/label=Cas9/note="generates RNA-guided double strand breaks in DNA"/translation="MDKKYSIGLDIGTNSVGWAVITDEYKVPSKKFKVLGNTDRHSIKKNLIGALLFDSGETAEATRLKRTARRRYTRRKNRICYLQEIFSNEMAKVDDSFFHRLEESFLVEEDKKHERHPIFGNIVDEVAYHEKYPTIYHLRKKLVDSTDKADLRLIYLALAHMIKFRGHFLIEGDLNPDNSDVDKLFIQLVQTYNQLFEENPINASGVDAKAILSARLSKSRRLENLIAQLPGEKKNGLFGNLIALSLGLTPNFKSNFDLAEDAKLQLSKDTYDDDLDNLLAQIGDQYADLFLAAKNLSDAILLSDILRVNTEITKAPLSASMIKRYDEHHQDLTLLKALVRQQLPEKYKEIFFDQSKNGYAGYIDGGASQEEFYKFIKPILEKMDGTEELLVKLNREDLLRKQRTFDNGSIPHQIHLGELHAILRRQEDFYPFLKDNREKIEKILTFRIPYYVGPLARGNSRFAWMTRKSEETITPWNFEEVVDKGASAQSFIERMTNFDKNLPNEKVLPKHSLLYEYFTVYNELTKVKYVTEGMRKPAFLSGEQKKAIVDLLFKTNRKVTVKQLKEDYFKKIECFDSVEISGVEDRFNASLGTYHDLLKIIKDKDFLDNEENEDILEDIVLTLTLFEDREMIEERLKTYAHLFDDKVMKQLKRRRYTGWGRLSRKLINGIRDKQSGKTILDFLKSDGFANRNFMQLIHDDSLTFKEDIQKAQVSGQGDSLHEHIANLAGSPAIKKGILQTVKVVDELVKVMGRHKPENIVIEMARENQTTQKGQKNSRERMKRIEEGIKELGSQILKEHPVENTQLQNEKLYLYYLQNGRDMYVDQELDINRLSDYDVDHIVPQSFLKDDSIDNKVLTRSDKNRGKSDNVPSEEVVKKMKNYWRQLLNAKLITQRKFDNLTKAERGGLSELDKAGFIKRQLVETRQITKHVAQILDSRMNTKYDENDKLIREVKVITLKSKLVSDFRKDFQFYKVREINNYHHAHDAYLNAVVGTALIKKYPKLESEFVYGDYKVYDVRKMIAKSEQEIGKATAKYFFYSNIMNFFKTEITLANGEIRKRPLIETNGETGEIVWDKGRDFATVRKVLSMPQVNIVKKTEVQTGGFSKESILPKRNSDKLIARKKDWDPKKYGGFDSPTVAYSVLVVAKVEKGKSKKLKSVKELLGITIMERSSFEKNPIDFLEAKGYKEVKKDLIIKLPKYSLFELENGRKRMLASAGELQKGNELALPSKYVNFLYLASHYEKLKGSPEDNEQKQLFVEQHKHYLDEIIEQISEFSKRVILADANLDKVLSAYNKHRDKPIREQAENIIHLFTLTNLGAPAAFKYFDTTIDRKRYTSTKEVLDATLIHQSITGLYETRIDLSQLGGD"CDS 8751..8771/codon_start=1/product="nuclear localization signal of SV40 (simian virus40) large T antigen"/label=SV40 NLS/translation="PKKKRKV"misc_feature 9521..9545/label=LB T-DNA repeat/note="left border repeat from nopaline C58 T-DNA"CDS 10073..10864/codon_start=1/gene="aadA"/product="aminoglycoside adenylyltransferase (Murphy,1985)"/label=Spe/note="confers resistance to spectinomycin andstreptomycin"/translation="MGEAVIAEVSTQLSEVVGVIERHLEPTLLAVHLYGSAVDGGLKPHSDIDLLVTVTVRLDETTRRALINDLLETSASPGESEILRAVEVTIVVHDDIIPWRYPAKRELQFGEWQRNDILAGIFEPATIDIDLAILLTKAREHSVALVGPAAEELFDPVPEQDLFEALNETLTLWNSPPDWAGDERNVVLTLSRIWYSAVTGKIAPKDVAADWAMERLPAQYQPVILEARQAYLGQEEDRLASRADQLEEFVHYVKGEITKVVGK"rep_origin 11108..11696/direction=RIGHT/label=ori/note="high-copy-number ColE1/pMB1/pBR322/pUC origin ofreplication"misc_feature 11882..12022/label=bom/note="basis of mobility region from pBR322"rep_origin 12366..12560/label=pVS1 oriV/note="origin of replication for the Pseudomonas plasmidpVS1 (Heeb et al., 2000)"CDS complement(12626..13693)/codon_start=1/product="replication protein from the Pseudomonas plasmidpVS1 (Heeb et al., 2000)"/label=pVS1 RepA/translation="GRKPSGPVQIGAALGDDLVEKLKAAQAAQRQRIEAEARPGESWQAAADRIRKESRQPPAAGAPSIRKPPKGDEQPDFFVPMLYDVGTRDSRSIMDVAVFRLSKRDRRAGEVIRYELPDGHVEVSAGPAGMASVWDYDLVLMAVSHLTESMNRYREGKGDKPGRVFRPHVADVLKFCRRADGGKQKDDLVETCIRLNTTHVAMQRTKKAKNGRLVTVSEGEALISRYKIVKSETGRPEYIEIELADWMYREITEGKNPDVLTVHPDYFLIDPGIGRFLYRLARRAAGKAEARWLFKTIYERSGSAGEFKKFCFTVRKLIGSNDLPEYDLKEEAGQAGPILVMRYRNLIEGEASAGS"CDS complement(14127..14756)/codon_start=1/product="stability protein from the Pseudomonas plasmidpVS1 (Heeb et al., 2000)"/label=pVS1 StaA/translation="MKVIAVLNQKGGSGKTTIATHLARALQLAGADVLLVDSDPQGSARDWAAVREDQPLTVVGIDRPTIDRDVKAIGRRDFVVIDGAPQAADLAVSAIKAADFVLIPVQPSPYDIWATADLVELVKQRIEVTDGRLQAAFVVSRAIKGTRIGGEVAEALAGYELPILESRITQRVSYPGTAAAGTTVLESEPEGDAAREVQALAAEIKSKLI"ORIGIN1 gtttacccgc caatatatcc tgtcaaacac tgatagttta aactagtgaa gtgaagcttg61 gtctcaacta tgagacctaa gctggcacaa ctatatttcc aacatcacta gctaccatca121 aaagattgac ttctcatctt actcgattga aaccaaatta acatagggtt tttatttaaa181 taaaagttta accttctttt taaaaaattg ttcatagtgt catgtcagaa caagagctac241 aaatcacaca tagcatgcat aagcggagct atgatgagtg gtattgtttt gttcgtcact301 tgtcactctt ttccaacaca taatcccgac aacaacgtaa gagcatctct ctctctccac361 acacactcat gcatgcatgc attcttacac gtgattgcca tgcaaatctc ctttctcacc421 tataaataca aaccaaccct tcactacact cttcactcaa accaaaacaa gaaaacatac481 acaaatagca aaacggtacc aacaatggat aacatggcca tcatcaagga gttcatgcgc541 ttcaaggtgc acatggaggg ctccgtgaac ggccacgagt tcgagatcga gggcgagggc601 gagggccgcc cctacgaggg cacccagacc gccaagctga aggtgaccaa gggtggcccc661 ctgcccttcg cctgggacat cctgtcccct cagttcatgt acggctccaa ggcctacgtg721 aagcaccccg ccgacatccc cgactacttg aagctgtcct tccccgaggg cttcaagtgg781 gagcgcgtga tgaacttcga ggacggcggc gtggtgaccg tgacccagga ctcctccctg841 caggacggcg agttcatcta caaggtgaag ctgcgcggca ccaacttccc ctccgacggc901 cccgtaatgc agaagaagac catgggctgg gaggcctcct ccgagcggat gtaccccgag961 gacggcgccc tgaagggcga gatcaagcag aggctgaagc tgaaggacgg cggccactac

